GlueX Technical Note
Request for Comments
HDDM - GlueX Data Model
draft 1.1
Richard Jones
September 12, 2003
(supersedes draft 1.0, September 12, 2003)
abstract
The GlueX experiment has adopted a policy that all shared data should
be described in xml. A set of tools has been developed that enable
programmers to respect this policy without the overhead of having
to do building and parsing of xml documents in user code. The data
are described in terms of data model structured like an xml document.
The general rules for building such model documents is called the
HDDM specification. Once a user has described the data model in xml,
tools are provided to translate the model into c structures, xml
schema, and a general-purpose i/o library.
Background
The workflow model for GlueX offline analysis is shown in Fig. 1.
It is important even at an early stage in the experiment to consider
carefully critical elements of the software framework in which these
components operate. One critical aspect of the framework is the way
the data are represented at intermediate stages in the pipeline. The
collaboration made an early decision to describe all such data in xml.
This choice brings with it the advantages of a host of automated tools
and libraries in the public domain for parsing and searching xml documents.
However it also demands careful design so that the additional software
layers introduced do not impose unintended restrictions on the client
programs.
A search on the web recently turned up a number of interesting projects
in which other groups are attempting to do the same or similar things.
Two projects stand out as most interesting from the point of view of
GlueX, the BinX project [1]
supported by the University of Edinburgh and the HFS project
[2] supported by NCSA, Urbana.
BinX is still in its infancy. None of the tools or libraries described
in the reference document appear to be available for downloading at the
time of of this writing. However from the description given, some
observations can be made. The aim is to provide a general metadata format
for describing just about any binary data file. The authors start from
the premise that storing and retrieving data files in xml format does not
make sense, and proceed to the conclusion that one never needs to actually
express them in xml. Thus their goal appears to be to use the xml as a
sort of documentation facility that is separate from the actual process
of handling and interpreting the data. This approach makes the most sense
when one is dealing with a set of i/o libraries and file formats that are
already in place, and wants to introduce a metadata description after the
fact. In GlueX we have the possibility of starting out using the xml in
the design process, and having the other pieces automatically created from
the xml. BinX adds the metadata as a new layer on top of, and independent
from, the layer in which programs actually process data, and as such it
looks more like a retrofit tool than an integrated model component.
However it is still new and seems to have some resources behind it, so
GlueX developers should follow it with interest.
The Hierarchical Data Format HDF is much older, by contrast, and seems to
already have established a base of scientific users. The design appears
to be oriented around the need to archive large multi-dimensioned arrays,
such as images. Its origins date back to the mid-1990's before the advent
of xml and reflects that fact in the frequent references to storing and
recovering data from fortran programs, but it is being actively developed
and in version 5 (HDF5) they have included new functionality to describe
data in xml. There are also java tools in the distribution, that probably
reflects the fact that support for scientific programming in java is on the
rise. The primary concern regarding the potential usefulness of HFS to
GlueX is the complexity of the data model and the program interface.
This package is designed to do much more than is needed by GlueX. For
example, the ability to delete arbitrary objects from existing data files,
or add new ones, complicates the i/o library considerably over what is
needed for HEP data streams. The number of operations that are required
to set up a new data file are considerable, compared with the simple
opens and writes to which HEP programmers are accustomed. From the point
of view of design, multi-dimensioned arrays are not the most appropriate
structure for expressing much of the content of a GlueX event. While it
could probably be made to work for GlueX, it appears that HDF would be
overkill for this problem. Thus there is still good reason to carry on
the earlier development of a xml data model for GlueX.
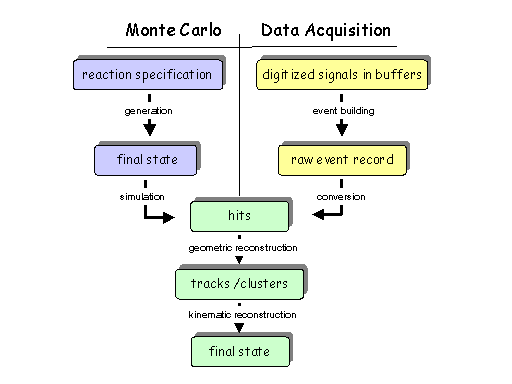 Fig. 1: The conceptual data model for GlueX begins with a physics
event, coming either from the detector or a Monte Carlo program,
which builds up internal structure as it flows through the
analysis pipeline. The data model specifies the elements of information
that are contained in a view of the event at each stage and the
relationships between them. The implementation provides standard
methods for creating, storing and accessing the information.
Fig. 1: The conceptual data model for GlueX begins with a physics
event, coming either from the detector or a Monte Carlo program,
which builds up internal structure as it flows through the
analysis pipeline. The data model specifies the elements of information
that are contained in a view of the event at each stage and the
relationships between them. The implementation provides standard
methods for creating, storing and accessing the information.
General Notes
- At each stage (lower-case items in diagram) in the pipeline one has
a unique view of the event.
- To each of these is associated a unique data model that expresses
the event in that view.
- GlueX policy is to use xml to describe all of our shared data, which
means any data that might be passed as input to a program or produced
as output. This does not mean that all data records are represented
as plain-text files, but that to each data file or i/o port is attached
some metadata that a tool can use to automatically express all of its
contents in the form of a plain-text xml document.
- This policy is interpreted to mean that to each data file or i/o port
of a program is associated a xml schema that defines the data structure
that the program expects or produces. The schemas should be either
bundled with the distribution of the program, or published on the web
and indicated by links in the Readme file.
- Any xml document should be accepted as input to a program if it is
valid according to the schema associated to that input port.
- In practice, this last requirement adds significant of overhead to the
task of writing a simple analysis program, because it must be capable of
parsing general xml documents as input. In addition to this overhead
imposed on the program code itself, the author must also produce schemas
for each input or output port or file accessed by the program.
- The purpose of the Hall D Data Model (HDDM) is to simplify the
programmer's task by providing automatic ways to generate schemas
and providing i/o libraries to standardize input and output of data
in the form of xml-described files.
- HDDM consists of a specification supported by a
set of tools to assist its implementation. The
specification is a set of rules that a programmer must obey in
constructing schemas in order for them to be supported by the tools.
The tools include an automatic schema generator and an i/o
library builder for c and c++.
- The HDDM specification was designed to enable the construction of an
efficient i/o library. It was assumed in the design that users could
not afford a general xml-parsing cycle every time an event is read in
or written out by a program. It was also assumed that serializing data
in plain-text xml is too expensive in terms of bandwidth and storage.
Using the HDDM tools, users can efficiently pass data between programs
in a serialized binary stream, and convert to/from plain-text xml
representations using a translation tool when desired.
- Programmers are not obligated to use HDDM tools to work in the GlueX
software framework. If they provide their own schema for each file or
i/o port used by the program and accept any xml that respects their
input schema then they are within the agreed framework.
- The HDDM tools are presently implemented in c and c++, so programmers
wishing to work in java have more work to do. However, they will find
it easy to interface to other programs that do use the HDDM libraries
because they provide for the correct reading and writing of valid xml
files and the automatic generation of schemas that describe them.
- The hddm-c tool automatically constructs a set of c-structures
based on xml metadata that can be used to express the data in memory.
It also builds an i/o library that can be called by user code to
serialize/deserialize these structures.
- The serialized data format supported by hddm-c consists of a
xml header in plain text that describes the structure and contents of
the file, followed by a byte string that is a reasonably compact
serialization of the structured data in binary form. Such hddm
files are inherently self-describing. The overhead of including the
metadata in the stream with the data is negligible.
- The hddm-xml tool extracts the xml metadata from a hddm file
header and expresses the data stored in the file in the form of a
plain-text xml document.
- The hddm-schema tool extracts the xml metadata from a hddm file
header and generates a schema that describes the structure of the data
in the file. The schema produced by hddm-schema will always
validate the document produced by hddm-xml when both act on the
same hddm file. More significantly, the schema can be used to check
the validity of other xml data that originate from a different source.
- The xml-hddm tool reads an xml document and examines its schema
for compliance with the HDDM specification. If successful, it parses
the xml file and converts it into hddm format.
- The schema-hddm tool reads a schema and checks it for compliance
with the HDDM specification. If successful, it parses it into the form
of a hddm file consisting of the header only and no data. Such a
data-less hddm file is also called a "template"
(see below).
- Note that the hddm-xml, hddm-schema, and hddm-c tools
can act on any hddm data file written by any program, even if the code
that produced the data is no longer available. This is because
sufficient metadata is provided in the schema header to completely
reconstruct the file's contents in xml, or instantiate it in c-structures.
- A tool called xml-xml has been included in the tool set as a
simple means to validate an arbitrary xml document against a dtd or
schema, and reformat it with indentation to make it easier to read.
- Tools called stdhep2hddm and hddm2stdhep provide
conversion between the hddm data stream and the STDHEP format used by
HDFast. This is an example where a user program achieves xml i/o
by employing translators, in this case a two-stage pipeline.
- In spite of the array of tools described above, the programmer still
must do the work of describing the structure and contents of the data
expected or produced by his program. He may do this in one of two
ways: either he constructs an original schema describing his data, or
he creates an original xml template of his data and then generates the
schema using hddm-schema.
- Since schemas are rather verbose and repetitive, the suggested method
is to create a template first, use hddm-schema to transform it
into a basic HDDM schema, and then add facets to the schema to enrich the
minimal set of metadata generated from the template. This method has
the advantage that one starts off with a basic schema that is known to
conform to the rules for HDDM schemas (see below)
so it is relatively simple thereafter to stay within the specification.
- As a shortcut to creating schemas, it is not necessary to do anything
more than just create the template. The basic schema that is generated
automatically from the template contains sufficient information to
validate most data, so a programmer can get by without ever learning
how to write or modify schemas.
Rules for constructing HDDM templates
- A hddm template is nothing more than a plain-text xml file that mimics
the structure of the xml that the program expects on input or produces
on output. In some ways it is like sample data that the programmer
might provide to a user to demonstrate how to use it, although the
comparison is not perfect.
- The top element in the template must be <HDDM> and have
three required attributes: class, version, and xmlns.
The value of the latter must be xmlns="http://www.gluex.org/hddm".
The values of the class and version arguments are user-defined. They
serve to identify a group of schemas that share a basic set of tags.
See below for more details on classes.
- The names of elements below the root <HDDM> element are
user-defined, but they must be constructed according to the following
rules.
- All values in hddm files are expressed as attributes of elements.
Any text that appears between tags in the template is treated as
a comment and ignored.
- An element may have two information attached to it: child elements
which appear as new tags enclosed between the open and close tags of
the parent element, and attributes which appear as key="value"
items inside the open tag.
- All quantities in the data model are carried by named attributes of
elements. The rest of the document exists to express the meaning of
the data and the relationships between them.
- All elements in the model document either hold attributes, contain other
elements, or both. Empty elements are meaningless, and are not allowed.
- One way a template is not like sample data is that it does not
contain actual numerical or symbolic values for the fields in the
structure. In the place of actual values, the types of the fields
are given. For example, instead of showing energy="12.5 as
might be shown for sample data, the template would show in this
position energy="float" or energy="double".
- The complete list of allowed types supported by hddm is "int", "long",
"float", "double", "boolean", "string", "anyURI", and "Particle_t". The
Particle_t type is a value from an enumerated list of capitalized
names of physical particles. The int type is a 32-bit signed integer,
and long is a 64-bit signed integer. The other cases are obvious.
- Attributes in the template that do not belong to this list are assumed
to be constants. Constants are sometimes useful for annotating the
xml record. The must have the same value for all instances of the
element throughout the template.
- Any given attribute may appear more than once throughout the template
hierarchy. Wherever it appears, it must appear with identical
attributes and with content elements of the same order and type.
- Another difference between a template sample data is that the
template never shows a given element more than once in a given context,
even if the given tag would normally the repeated more than once for
an actual sample. One obvious example of this is a physics event,
which is represented only once in the template, but repeated multiple
times in a file.
- By default, it is assumed that an element appearing in the template
must appear in that position exactly once. If the element is allowed
to appear more than once or not at all then additional attributes
should be inserted in the element of the form minOccurs="N1"
and maxOccurs="N2", where N1 can be zero or any positive
integer and N2 can be any integer no smaller than N1, or
set to the string "unbounded". Each defaults to 1.
- Arrays of simple types are represented by a sequence of elements,
each carrying an attribute containing a single value from the array.
This is more verbose than allowing users to include arrays as a simple
space separated string of values, but the chosen method is more apt
for expressing parallelism between related arrays of data.
- An element may be used more than once in the model, but it may never
appear as a descendent of itself. Such recursion is complicated to
handle and it is hard to think of a situation where it is necessary.
- Examples of valid hddm templates are given in the examples section
below.
- Because templates contain new tags that are invented by the programmer,
it is not possible to write a standard template schema against which a
programmer can check his new xml file for use as a template. Instead of
using schema validation, the programmer can use the hddm-schema
tool to check a xml file for correctness as a hddm template. Any errors
that occur in the hddm - schema transformation indicate problems in the
xml file that must be fixed before it can be used as a template.
Rules for constructing HDDM schemas
- HDDM schemas must be valid xml schemas, belonging to the namespace
http://www.w3.org/2001/XMLSchema. Not every valid schema is a valid
HDDM schema, however, because xml allows for several different ways to
express a given data structure.
- GlueX programmers are not obligated to write schemas that conform to
the HDDM specification, but if they do, they have the help of the HDDM
tools for efficient file storage and i/o.
- In the following specification, a prefix xs: is applied to the
names of elements, attributes or datatypes that belong to the official
schema namespace "http://www.w3.org/2001/XMLSchema", whose meaning is
defined by the xml schema standard. The extensions introduced for the
specific needs of GlueX are assigned to a private namespace called
"http://www.gluex.org/hddm" that is denoted by the prefix hddm:.
- The top element defined by the schema must be <hddm:HDDM> and have
three required attributes: class, version, and xmlns.
The value of the latter must be xmlns="http://www.gluex.org/hddm".
The class and version arguments are of type xs:string and are
user-defined. They serve to identify a group of schemas that share a
basic set of tags. See below for more details.
- The names of elements below the root <hddm:HDDM> element are
user-defined, but they must be constructed according to the following
rules.
- An element may have two kinds of content: child elements and attributes,
and hence must have xs:complexType. Elements represent the
grouping together of related pieces of data in a hierarchy of nodes.
The actual numerical or symbolic values of individual variables appear
as the values of attributes. Examples are shown
below.
- All quantities in the data model are carried by named attributes of
elements. The rest of the document exists to express the meaning of
the data and the relationships between them.
- All elements in the model document either hold attributes, contain other
elements, or both. Empty nodes are meaningless, and are not allowed.
- Text content between open and close tags is allowed in documents
(type="mixed") but it is treated as a comment and stripped on
translation. Basic HDDM schemas do not use type="mixed"
elements.
- The datatype of an attribute is restricted to a subset of basic types
to simplify the task of translation. Currently the list is
xs:int, xs:long, xs:float, xs:double,
xs:boolean, xs:string, xs:anyURI and
hddm:Particle_t. User types that are derived from the above
by xs:restriction may also be defined and used in a HDDM schema.
- Attributes must always be either "required" or "fixed". Default
attributes, i.e. those that are sometimes present inside their host and
sometimes not are not allowed. This allows a single element to be
treated as a fixed-length binary object on serialization, which has
advantages for efficient i/o.
- A datum that is sometimes absent can be expressed in the model by
assigning it as an attribute to its own host element and putting the
host element into its parent with minOccurs="0".
- Fixed attributes (with use="fixed") may be attached to
user-defined elements. They may be of any valid schema datatype, not
just those listed above, and may be used as comments to qualify the
information contained in the element. Because they have the same
value for every instance of the element, they do not take up space in
the binary stream, but they are included explicitly in the output
produced by the hddm-xml translator.
- All elements must be globally defined in the schema, i.e. declared at
the top level of the xs:schema element. Child elements are
included in the definition of their parents through a ref=tagname
reference. Local definitions of elements inside other elements are not
allowed. This guarantees that a given element has the same meaning and
contents wherever it appears in the hierarchy.
- Arrays of simple types are represented by a sequence of elements,
each carrying an attribute containing a single value from the array.
This is more verbose than allowing a simple list type like is defined
by xs:list, but the chosen method is more apt for expressing
parallelism between related arrays of data, such as frequently occurs
in descriptions of physical events. Forbidding the use of simple
xs:list datatypes should encourage programmers to chose the
better model, although of course they could just mimic the habitual use
of lists by filling the data tree with long strings of monads!
- Elements are included inside their parent elements within a
xs:sequence schema declaration. Each member of the sequence
must be a reference to another element with a top-level definition.
- A given element may occur only once in a given the sequence, but may
have minOccurs and maxOccurs attributes to indicate
possible absence or repetition of the element.
- The sequence is the only content organizer allowed by HDDM.
More complex organizers are supported by schema standards, such as
all and choice, but their use would complicate the i/o
i/o interfaces that have to handle them and they add little by way
of flexibility to the model the way it is currently defined.
- An element may be used more than once in the model, but it may never
appear as a descendent of itself. Such recursion is complicated to
handle and it is hard to think of a situation where it is necessary.
- A user can check whether a given schema conforms to the HDDM rules
by transforming it into a hddm template
document. Any errors that occur during the transformation generate
a message indicating where the specification has been violated.
Class relationships between HDDM schemas
- Two HDDM schemas belong to the same class if all tags that are
defined in both have the same set of attributes in both.
- This is a fairly weak condition. It is possible that all data files
used in GlueX will belong to the same class, but it is not required.
- If two HDDM schemas belong to the same class then it is possible to
form a union schema that will validate documents of either type by
taking the xml union of the two schema documents and changing any
sequence elements in one and not in the other to minOccurs="0".
- The translation tools xml-hddm and hddm-xml will work
with any HDDM class.
- Any program built using the i/o library created with hddm-c is
dependent on the class of the schema used during the build. Any files
it writes through this interface will be built on this schema, however
it is able to read any file of the same class without recompilation.
- A new schema may be derived from an existing HDDM schema by taking the
existing one and adding new elements to the structure. In this case
the version attribute of the HDDM tag should be incremented, while
leaving the class attribute unchanged.
- A program that was built using the hddm-c tool for its i/o
interface can read from any from any hddm file of the same class as
the original schema used during the build. If the content of the file
is a superset of the original schema then nothing has changed. If
some elements of the original schema are missing in the file then the
i/o still works transparently, but the c-structures corresponding to
the missing elements will be empty, i.e. zeroed out.
- The c/c++ i/o library rejects an attempt to read from a hddm file that
has a schema of a different class from the one for which it was built.
- No mandatory rules are enforced on the version attribute of the
hddm file, but it is available to programs and may be used to select
certain actions based on the "vintage" of the data.
- Programs that need simultaneous access to multiple classes of hddm
files can be built with more than one i/o library. The structures and
i/o interface are defined in separate header files hddm_X.h and
implementation files hddm_X.c, where X is the class letter.
Implementation Notes
- There is a complementarity between xml schemas and the xml templates
that express the metadata in hddm files. Depending on the level of
detail desired, schemas may become arbitrarily sophisticated and
complex. On the other hand, only a small subset of that information
is needed to support the functions of the hddm tool set. Templates
allow that information to be distilled in a compact form that is both
human-readable and valid xml.
- In the present implementation, the text layout of the template
(including the whitespace between the tags) is used by the hddm tools
to simplify the encoding and decoding. There is exactly one tag per
line and two leading spaces per level of indent. This may change in
future implementations. This means that hddm file headers should not
be edited by hand.
- The XDR [3]
library is used to encode the binary values in the hddm
stream. This means that hddm files are machine-independent, and
can be read and written on any machine without any dependence on
whether the machine is little-endian or big-endian. XDR is the network
encoding standard library developed for Sun's rpc and nfs services.
For more info, search for RFC 1014 on the web or do "man xdr" under
linux.
- The binary file format will change. The point is not to fix
on some absolute binary format at this early stage. The only
design constraint was that the data model be specified in xml and
that the data be readily converted into plain-text xml, preferably
without needing to look up auxiliary files or loading the libraries
that wrote it.
- The design of the i/o library has been optimized for flexibility:
the user can request only the part of the model that is of interest.
The entire model does not even have to be present in the file, in which
case only the parts of the tree that are present in the file are loaded
into memory, and the rest of the requested structure is zeroed out.
- The only constraint between the model used in the program and that
of the hddm stream is that there be no collisions, that is tags
found in both but with different attributes.
- Two data models with colliding definitions can be used in one program
but they have to have different class Ids. Two streams with
different class Ids cannot feed into each other. In any case the
xml viewing tool hddm-xml can read a hddm stream of any class.
Examples
- A simple model of an event fragment describing hits in a
time-of-flight wall. It allows for multiple hits per detector
in a single event, with t and dE information for each hit.
The hits are ordered by side (right: end=0, left: end=1) and then by
horizontal slab. The minOccurs and maxOccurs attributes allow those
tags to appear any number of times, or not at all, in the given context.
<forwardTOF>
<slab y="float" minOccurs="0" maxOccurs="unbounded">
<side end="int" minOccurs="0" maxOccurs="unbounded">
<hit t="float" dE="float" maxOccurs="unbounded" />
</side>
</slab>
</forwardTOF>
- A model of the output from an event generator.
An example of actual output from genr8
converted to xml using hddm-xml. Warning: some browsers have
difficulty displaying plain xml. Mozilla 1.x and Internet Explorer 6
give a nice view of the document below.
<?xml version="1.0" encoding="UTF-8"?>
<HDDM class="s" version="1.0" xmlns="http://www.gluex.org/hddm">
<physicsEvent eventNo="int" runNo="int">
<reaction type="int" weight="float" maxOccurs="unbounded">
<beam type="Particle_t">
<momentum px="float" py="float" pz="float" E="float" />
<properties charge="int" mass="float" />
</beam>
<target type="Particle_t">
<momentum px="float" py="float" pz="float" E="float" />
<properties charge="int" mass="float" />
</target>
<vertex maxOccurs="unbounded">
<product type="Particle_t" decayVertex="int" maxOccurs="unbounded">
<momentum px="float" py="float" pz="float" E="float" />
<properties charge="int" mass="float" />
</product>
<origin vx="float" vy="float" vz="float" t="float" />
</vertex>
</reaction>
</physicsEvent>
</HDDM>
- A more complex example follows, showing a hits tree for the full
detector.
<?xml version="1.0" encoding="UTF-8"?>
<HDDM class="s" version="1.0" xmlns="http://www.gluex.org/hddm">
<physicsEvent eventNo="int" runNo="int">
<hitView version="1.0">
<barrelDC>
<cathodeCyl radius="float" minOccurs="0" maxOccurs="unbounded">
<strip sector="int" z="float" minOccurs="0" maxOccurs="unbounded">
<hit t="float" dE="float" maxOccurs="unbounded" />
</strip>
</cathodeCyl>
<ring radius="float" minOccurs="0" maxOccurs="unbounded">
<straw phim="float" minOccurs="0" maxOccurs="unbounded">
<hit t="float" dE="float" minOccurs="0" maxOccurs="unbounded" />
<point z="float" dEdx="float" phi="float"
dradius="float" maxOccurs="unbounded" />
</straw>
</ring>
</barrelDC>
<forwardDC>
<package pack="int" minOccurs="0" maxOccurs="unbounded">
<chamber module="int" minOccurs="0" maxOccurs="unbounded">
<cathodePlane layer="int" u="float" minOccurs="0" maxOccurs="unbounded">
<hit t="float" dE="float" minOccurs="0" maxOccurs="unbounded"/>
<cross v="float" z="float" tau="float" maxOccurs="unbounded" />
</cathodePlane>
</chamber>
</package>
</forwardDC>
<startCntr>
<sector sector="float" minOccurs="0" maxOccurs="unbounded">
<hit t="float" dE="float" maxOccurs="unbounded" />
</sector>
</startCntr>
<barrelCal>
<module sector="float" minOccurs="0" maxOccurs="unbounded">
<flash t="float" pe="float" maxOccurs="unbounded" />
</module>
</barrelCal>
<Cerenkov>
<module sector="float" minOccurs="0" maxOccurs="unbounded">
<flash t="float" pe="float" maxOccurs="unbounded" />
</module>
</Cerenkov>
<forwardTOF>
<slab y="float" minOccurs="0" maxOccurs="unbounded">
<side end="int" minOccurs="0" maxOccurs="unbounded">
<hit t="float" dE="float" maxOccurs="unbounded" />
</side>
</slab>
</forwardTOF>
<forwardEMcal>
<row row="int" minOccurs="0" maxOccurs="unbounded">
<column col="int" minOccurs="0" maxOccurs="unbounded">
<flash t="float" pe="float" maxOccurs="unbounded" />
</column>
</row>
</forwardEMcal>
</hitView>
</physicsEvent>
</HDDM>
References
[1]
Representing Scientific Data on the Grid with BinX, Binary XML Description
Language, M. Westhead and M. Bull, University of Edinburgh, January 2003.
[2]
HDF5 Users's Guide,
working draft, July 2003.
[3]
RFC 1832 (rfc1832) -
XDR: External Data Representation standard, September 1995.
This material is based upon work supported by the National Science Foundation under Grant No. 0901016.

